In 2005, as high throughput sequencers became commercially available, the largest nucleotide sequence databases in the world joined forces to create the International Nucleotide Sequence Database (INSD). Today, the European, American, and Japanese archives exchange information daily, and the INSD ensures that nucleotide sequence data and its corresponding metadata are preserved as part of the scientific record, for future generations to reuse.
Since its creation, the number of nucleotide sequences in the INSD has grown exponentially. But how much of this data is really reusable? We began to ask ourselves this question when we set out to collect bacterial community data for meta-analyses. While the archiving of ecological data is the subject of ongoing discussion, we expected microbial data to be more accessible: their format is generally homogeneous (i.e., nucleotide sequence reads), data archiving has been centralized for over a decade, and journals in microbial ecology have implemented increasingly stringent and precise data deposition guidelines.
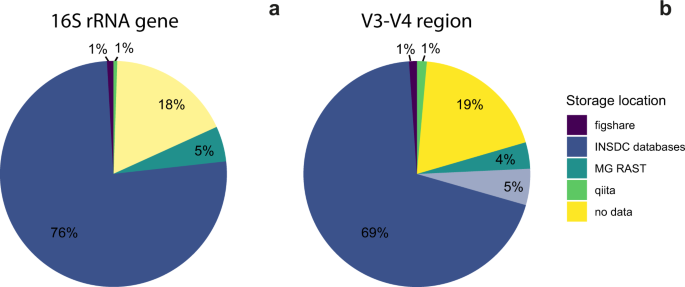
However, this was not the case—we encountered problems with approximately half of the datasets. We wanted to know why these issues arose, but this is experimentally hard to determine. In initial surveys, we had found that articles often deposited their nucleotide sequence data to the INSD, but minor errors downstream (i.e., archiving and documenting the data and metadata) rendered the sequences either inaccessible or not reusable. So instead, we decided to ask: where in the data deposition process do nucleotide sequence data stop being reusable, and with which frequency does each error occur?
We divided the data archiving process into four criteria which were necessary for data reuse (data location, deposition, formatting, and labeling). We examined datasets from articles published in microbial ecology-specific journals, as we had found that these journals had more precise requirements for data deposition. Then, we checked if the data met these four criteria.
Our findings shed some light into why data gets lost. One reason is the rapid pace with which sequencing technologies and best practices change, which make it hard to preserve all the data which is necessary to reanalyze the sequences in the future. One example is the frequent lack of mapping files, which are required to demultiplex sequence files into individual samples.
Our study also highlights that the solution may require little additional effort, but entails data providers, databases, and journals working together more closely. Even when data providers had uploaded their sequences to an INSDC database and had provided accession numbers, these accession numbers were often incorrect, or the data had not been made public—additional checking that the accession numbers are correct may be a simple and effective way that data does not get lost. Sending reminders to data providers to make their data public upon article publication may be another.
As the popularity of nucleotide sequencing continues to grow, so will the databases where these data are archived. Ensuring that the archives remain full of reusable data is an investment in the future of our field.







Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in