In 2009, the UK Medical Research Council launched a highly insightful call for proposals called ESEI – Environmental and Social Ecology of Human Infectious Diseases, which aimed to shed light on how a change in our natural, physical and social environments through globalisation, urbanisation, deforestation, changing food production and lifestyles may be inadvertently altering the patterns of disease spread. Some years down the line, the report in Nature Microbiology today is part of the result of that story. Here, we tell the back story to the work in the field in Kenya that led to this paper.
The context
Today, more than half of the human race lives in cities, and this number is set to continue increasing. In Africa, a combination of growth and migration is resulting in substantial increases in the population of urban and peri-urban zones, rising from 35% of the total population in 2007 to a projected 51% by 2030. Urbanised environments in Africa are melting pots of activity and interaction: the wealthy live alongside the poor; livestock live alongside people; human and livestock waste is poorly disposed of near food production areas; formal and informal trade takes place in internal and externally connected networks. This degree of mixing and contact creates ecological niches with opportunities for pathogen transmission. Indeed, a number of influential reports in the early 2000s linked urbanization to the risk of emerging infectious diseases (EIDs). As has become abundantly clear since 2019, EIDs are a major concern to the global public health community (and to the people of the world in general), and understanding the processes that lead to emergence is therefore a scientific research priority.
.jpg) |
.jpg) |
Images of low income parts of Nairobi where parts of our study took place
Approximately 60% of human pathogens are zoonotic, and approximately 80% of novel pathogens have zoonotic origins. The interface between livestock and humans is a potentially important source of pathogen emergence into humans: 40% of known livestock pathogens (>200 species) can infect humans. Further, almost all cities globally host extensive populations of human-adapted wildlife, be that foxes, rats, hyaenas or sparrows, a fact that is particularly noticeable in the tropics where cities frequently sit astride zones of high biodiversity. These urban-adapted species tend to harbour more known pathogens, making them important components of the human-animal interface in cities.

Chickens - a very popular livestock species in dense urban settings
What did we do?
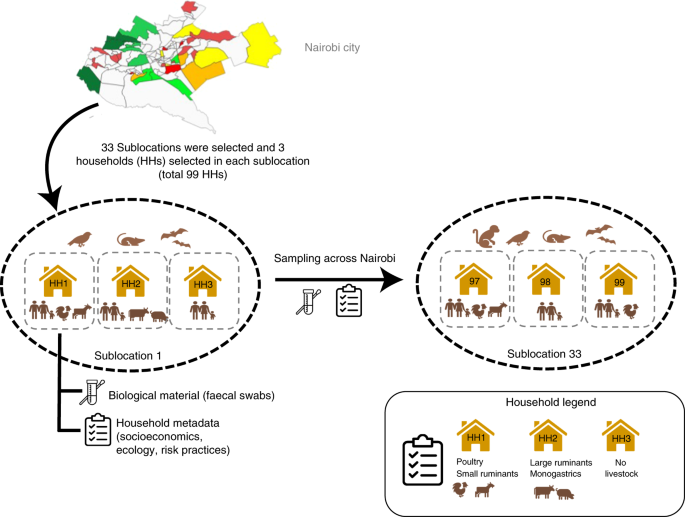
We adopted a landscape genomics approach to understanding the connectivity between populations in a complex urban setting. We chose Nairobi, Kenya (see map) as our study site because it is growing fast, is internationally connected, and riddled with interfaces between different habitat types. Kenya also has a fantastic scientific workforce and respected institutions in public health, demography, microbiology, livestock health and ecology, making collaborative networks strong and fruitful. Nairobi currently covers 690km2 and sustains 4.5 million people. 50% of households in some parts of the city keep livestock. Informal settlements – slums – represent up to 60-70% of the city’s population, similarly to other fast-growing African cities.

The Urban Zoo project, as we called it, was interdisciplinary – there were many components led by specialists in their field, including epidemiologists, public health and veterinary public health specialists, urban planners, agricultural economists, natural resource specialists, sociologists, geographers, ecologists, microbiologists and microbial geneticists and demographers. Each component informed the way the other components were operationalised. For example, we spent 3 years studying the structure of all the animal source food value chains in Nairobi, building a detailed picture of how live animals and their products flow into the city. We studied the urban planning history and contemporary planning decisions, focussing on the city planners’ often conflicting views of urban livestock keeping. We mapped social and economic gradients in the city, conducted detailed ecological assessments of micro-scale interfaces between different species and conducted a large-scale study of diarrhoea in children in slums. A core part of what we did was called the 99-households study, and it is about that part of this huge project that we are reporting in Nature Microbiology today.

The Urban Zoo 99 household team, at the International Livestock Research Institute, in Nairobi, Kenya
We used Escherichia coli as an exemplar organism to understand the connectivity between populations through the ‘eyes’ of a micro-organism. This bacterium is ideal because it is zoonotic, it exists in many hosts, on animal source foods and in most environments. Some E. coli strains are pathogens, but more broadly, E. coli is a commensal organism that is ubiquitous in microbial flora. Since many researchers before us have developed the genetic tools to study the diversity of E. coli, we could lean on work that had come before rather than develop an entirely new toolbox. A primary aim was to understand urban livestock keeping, and consumption of more distantly produced livestock products, as a driver of emerging infectious disease.
We collected around 2100 highly structured samples from livestock, humans, wildlife and the environment, following a true One Health approach, in our 99 participating households. This translated to just under 10,000 petri dishes for microbial culture and 12,500 cryovials of frozen samples that are now securely biobanked in Nairobi. Doing this sampling in a busy city over a period longer than a year, was not without complexities. Firstly, microbiology elements were led by two different institutional teams – the University of Nairobi’s Veterinary Public Health Pharmacology & Toxicology department and the Kenya Medical Research Institute’s Centre for Microbiology Research.
Both labs ran the same protocols, conducted cross-lab quality control, and processed field samples the same day, so the timing was everything to get fresh material to the bench, through the traffic – on the back of our subcontracted motorbikes.

Sample transfer in a busy city is best done by motorbike
Fieldwork was conducted by multiple, co-ordinated teams, working in 2 households per fieldwork day. Catching birds required starting at 4am – so as to put mist nets up before the sun rises at 0630 (at the same time every day, in this city just 2° below the Equator).

To catch bats, the same team, having checked rodent traps, processed samples in the lab and then had a rest, had to work through dusk and into the early night.

Meanwhile, a household team, which had recruited each household, secured informed consent, and dealt with human and animal sampling ethics, visited the house mid-morning to sample the people, their livestock and the broader environment.

One Health in action - sampling humans, livestock, wildlife and the environment within and around households
The project brought a team of medical and veterinary professionals to the households, and once lab work was complete and preliminary results generated, a comprehensive report was fed back to members of each household, with any referral or treatment advice given confidentially.
Fieldwork was punctuated with the occasional security incident (some neighbourhoods were a bit sketchy, particularly at night), refusal to participate (not as frequent as one might think), vehicle breakdowns, supply chain disruptions, Presidential elections (three of them!), and other logistic elements. Coordination of each team, the teams overall, and the quality control coordination of the labs, was an essential effort.
Each laboratory conducted a full microbiological work-up on each sample (including AST – phenotypic antimicrobial susceptibility testing), engaging multiple students to learn microbiological techniques through that effort. When the project started, the costs of tools to characterise the molecular diversity of the E. coli isolates was a major concern. At the very beginning, it seemed that the cost of whole-genome sequencing on this scale would be prohibitive, and samples in the field were randomly allocated ‘red dots’ (see picture) to denote which ones should go for sequencing. As the project proceeded, the costs and availability of sequencing changed though, and eventually, an isolate from each and every one of the hosts sampled was sequenced (and in some cases, up to 10 per host - see supplementary information in the paper).
 |
 |
The findings
Bacterial diversity at the scale of a city is extensive: effectively, the city is a 'soup' within which much mixing occurs, and within which the global diversity of E. coli can be found.
There are few host boundaries limiting this diversity: isolates were found to be shared by humans and between human and animal populations.
The household, our primary unit of analysis in this study, was very relevant: within a household, extensive sharing between hosts takes place, but the household is a hard nut to crack for a new strain, with the household environment selecting what gets admitted to that micro-environment. This is very relevant epidemiologically - we often discuss transmission using the analogy of a wave, but in fact it's a story of multiple short infections or transmission events that then lead to a multitude of infections within those micro populations. It's not moving as a wave, but through a much more structured series of events. This understanding of structuring helps us formulate policy that protects the health of people, livestock and wildlife in cities. Practically, our findings from the 99-household study provide insight into where surveillance for animal-borne emerging pathogens should be targeted, and could be used to explore pre-emptive measures that disrupt animal-to-human spillover – such as better management of animal waste.







Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in