The treatment of wastewater is one of the largest bio-technological processes in the world. Aeration tanks use an extremely nutrient rich environment to sustain a live and bustling community of microbes that break down organic matter. These microbial communities suspended in wastewater are called activated sludge and the process creates visible flocs of microbes that settle to the bottom of an aeration tank, allowing the clearer water to flow away for further treatment.
My interest in activated sludge begun while I was a student at Queen Mary University of London. I was encouraged by my MSc supervisor Pavel Kratina, a population and community ecologist. Pavel and his successful research group seek to understand the mechanistic processes that drive species diversity and control ecosystem functioning. While I was investigating potential research topics, Pavel introduced me to Alfred Burian, a post-doc researcher from Derby University. Alfred was recruiting a research partner to explore how environmental stress in planktonic communities may have knock-on effects on their services. We needed a hyper-diverse and productive microbial community to test our ecological concepts and activated sludge ticked all our boxes. They are dense microbial communities sustained in nutrient rich environments, susceptible to huge shifts in community composition and provide an essential service to humanity by cleaning wastewater.
 Photo 1: Alfred Burian at Derby Sewage Treatment Plant overlooking the aeration tanks
Photo 1: Alfred Burian at Derby Sewage Treatment Plant overlooking the aeration tanks
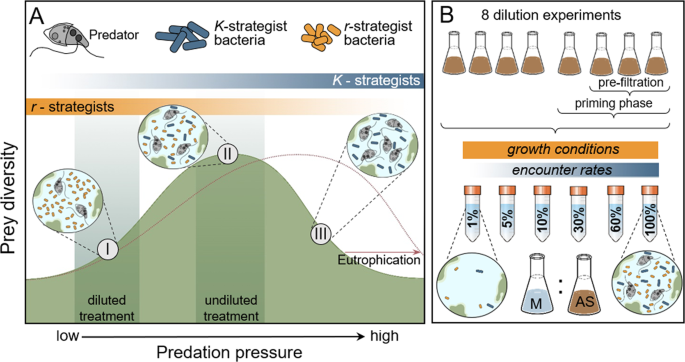
Activated sludge communities provide us with an insight on how disturbances alter natural ecosystem and what mechanisms maintain high diversity in many places of our planet. A key ecological theory posits that high diversity in highly productive ecosystems (such as activated sludge) can be maintained by high levels of predation pressure. As such, my collaborators and I set on a quest to test this theory experimentally in activated sludge communities. More specifically, we manipulated the predation pressure that protozoans (single-cell predators that feed on bacteria) impose on microbial communities. We predicted that increased predation would allow co-existence of more diverse forms of microbes, many of which would be outcompeted by a few fast-growing bacterial groups in the systems that lack or have only low levels of predation.
I was inspired by classic experimental work from Landry and Hassett in 1982, who reduced grazing pressures of marine plankton through diluting their environment (saltwater). By diluting the environment inhabited by both predators and their prey (by adding water), one could reduce the rate at which these organisms encounter each other, ultimately reducing predation pressure. We mirrored this experimental design and set up a series of laboratory dilutions of the replicated activated sludge communities, to generate an experimental gradient from very low to very high pressure of protozoan predation.
With the help of our site contact Peter Vale, we captured our activated sludge from operational aeration tanks at Severn Water treatment plant in Derby. Our sludge was then driven in the back of a car boot to Derby University where it was kept safe from humans in a make-shift biohazard zone. To keep our microbial communities alive, we nurtured them in a series of glass containers called chemostats– see Figure 1. The chemostats enabled us to simulate the conditions of an aeration tank on a miniaturised scale by continuously adding fresh media and removing waste culture. These artificial and simplified ecosystems were designed so that our predators could catch their prey under a constant resource level. Our mini eco-systems bubbled within their biohazard zone for two weeks as part of the conditioning phase, allowing us to keep environmental conditions constant. By doing this we could set-up different predator assemblages prior to our dilution experiment.

Figure 1: Picture of our four chemostats set at different dilution rates to condition our microbial communities.
After this conditioning phase, I performed a series of dilution experiments and sampled communities across a gradient of predation pressure. The samples were first analysed using flow cytometry, a technique commonly used in medicine to count microscopic cell populations by detecting their light scatter from a laser beam. I used a flow cytometer at the University of Roehampton to measure the population sizes of protists and bacteria in our experimental sludge communities. This was completed with the expertise of Ignacio Peralta-Maraver, who guided me through the flow cytometric know-how. Next-generation sequencing was then used in tandem with flow cytometry to observe changes in multiple aspects of bacterial diversity and community composition.

Photo 2: Me and Ignacio setting up samples for flow cytometric analysis at the University of Roehampton
We found that protozoan predation profoundly impacted bacterial diversity and community composition with potentially far-reaching implications for wastewater treatment (Burian et al. 2021). Both the decrease of prey encounter rates through dilution and the removal of top predators via filtration substantially reduced alpha, beta and phylogenetic diversity of microbes. Reducing predation also led to the decline of multiple groups of bacteria (Comamonadaceae, Nitrospira and Candidatus Accumulibacter) that support wastewater treatment efficiency via processes such as nitrogen removal. Although we have not measured the changes in ecosystem functions directly, these findings lend a strong support to our thesis that strong predation by protozoans is essential in the efficiency of this important bio-technological processes.
Video 1: Sludge flocs in our experimentally diluted microcosm.
The positive effect of predation on prey diversity is likely controlled by the selective feeding of protozoans on faster-growing bacteria that have not invested in defensive strategies. This agrees with so called ‘Kill the Winner’ hypothesis which explains how predation maintains diversity through reducing the dominance of strong and fast-growing competitors. However, bacterial diversity also suffered under very high predation pressures suggesting a hump-shaped relationship between predation rate and prey diversity in our activated sludge communities. This may be because protozoans account for very high proportion of in activated sludge biomass, and extremely high predation may negate their beneficial effects on prey diversity.
Video 2: Bubbling chemostats running at different dilution rates to condition our sludge communities.
After completing my research, I was driven to continue working with the water industry. This led to me finding employment as a microbiologist for Thames Water. Looking forward, the water industry is looking to better measure and develop their knowledge on the essential functions micro-organisms can provide treatment processes. For example, flow-cytometric instruments are now being considered for use as a potential tool for measuring water quality. There are huge benefits to be gained from considering the very tiny and often overlooked protozoa in microbial communities as a tool to both assess and control wastewater treatment. Research such as ours (Burian et al. 2021) represent a crucial step forward in advancing general ecological theory as well as improving the capacity of biological treatments in the activated sludge process.

Photo 3: Me working as a Microbiologist for Thames Water after finishing my MSc at Queen Mary University of London.
References:
Burian, A., Pinn, D., Peralta-Maraver, I., Sweet, M., Mauvisseau, Q., Eyice, O., Bulling, M., Röthig, T. and Kratina, P. 2022. Predation increases multiple components of microbial diversity in activated sludge communities. The ISME Journal, https://doi.org/10.1038/s41396-021-01145-z
Landry, M.R., Hassett, R.P. 1982. Estimating the grazing impact of marine micro-zooplankton. Marine Biology 67, 283–288. https://doi.org/10.1007/BF00397668






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in