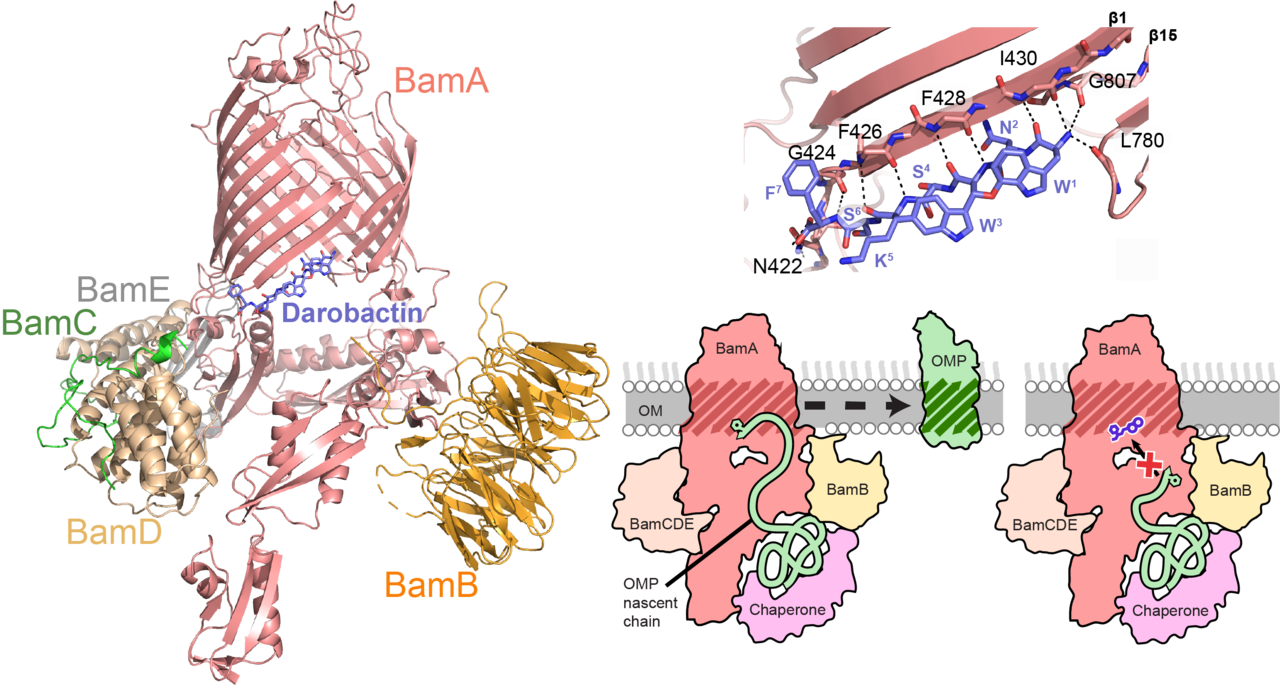
Bacteria are becoming increasingly resistant to antibiotics and there has been no new class of antibiotics in 30 years (1,2,3). The problem is especially grave for Gram-negative bacteria that have an additional outer membrane protecting the cell against xenobiotics. Consequently, essential proteins in the outer membrane are attractive antibiotic targets. One such target is the essential β-barrel assembly machinery (BAM complex). This 5-protein complex that is a “membrane chaperone” inserts the outer membrane β-barrel proteins (OMPs) into the outer membrane (4,5). Very recently, darobactin, a ribosomally synthesized and post-translationally modified heptapeptide was identified from a screen of Photorhabdus strains (6). Darobactin is highly active against clinically relevant Gram-negative pathogens, however, its size makes it too big to penetrate across the bacterial outer membrane. There were two main things that fascinated us: Firstly, direct interaction studies showed that it targets BamA, which does not have a catalytic center. Secondly, what is the “warhead” of the antibiotic molecule darobactin? Is it the two intricately locked rings or the peptide backbone? In order answer these, we looked into the structural aspects of BamA-darobactin interaction.
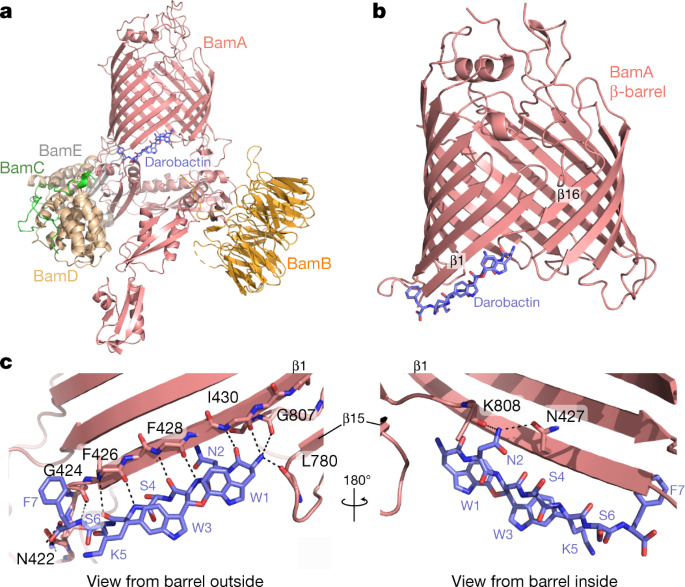
For our work, we integrated three structural techniques - NMR spectroscopy, X-ray crystallography and cryo-electron microscopy (cryo-EM). Solution NMR spectroscopy on the BamA-β barrel showed that darobactin stabilizes BamA in a single conformation, which otherwise oscillates between two states. Cryo-EM of the BAM complex and X-Ray crystallography on the BamA-β barrel showed one molecule of darobactin bound to BamA at a unique position in the “gate region”. This region is located between the first and the last strand of the barrel, where a lipid molecule has been observed bound in previously reported studies. Thus, we identified a highly unconventional binding site at the interface of water, protein and membranes. Using molecular dynamics (MD) simulations and native mass spectrometry, we found out that due to its special bicyclic structure, darobactin is a ready-to-bind molecule that perfectly fits the unique environment at the gate region. Surprisingly, the peptide backbone is the “warhead” of darobactin as it binds to the open gate of BamA. This distinct feature that most of the contacts between BamA and darobactin are backbone hydrogen-bonds, makes the interaction robust against resistance by mutagenesis. We have verified this by isothermal titration calorimetry (ITC) binding studies with alanine mutants of the darobactin binding site on BamA.

Remarkably, darobactin resembles the signal sequences that most BAM substrates contain at their C-termini, for insertion into the outer membrane. We carried out competition binding experiments between darobactin and β-signal peptides from different OMPs and saw that darobactin outcompetes the peptides. The bicyclic structure rigidifies the peptide backbone, making it a perfect match for binding to open gate form of BamA, thus blocking it from interacting with any other linear peptide. It thus appears that darobactin blocks the first step of substrate insertion catalyzed by BamA, leading to cell death.
The full article contains all details about the experiments mentioned here, as well as additional mechanistic and microbiological insights:
https://www.nature.com/articles/s41586-021-03455-w.pdf
References:
1 Brown, E. D. & Wright, G. D. Antibacterial drug discovery in the resistance era. Nature 529, 336-343, (2016).
2 Lewis, K. The science of antibiotic discovery. Cell 181, 29-45, (2020).
3 Tacconelli, E. et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18, 318-327, (2018).
4 Bakelar, J., Buchanan, S. K. & Noinaj, N. The structure of the β-barrel assembly machinery complex. Science351, 180-186, (2016).
5 Gu, Y. et al. Structural basis of outer membrane protein insertion by the BAM complex. Nature 531, 64-69, (2016).
6 Imai, Y. et al. A new antibiotic selectively kills Gram-negative pathogens. Nature 576, 459-464, (2019).






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in