Banner photo credit: Anna Roik, GEOMAR Helmholtz Centre for Ocean Research Kiel

I study coral reefs for the same reasons they captive the imaginations of most. The landscapes are breathtaking and the animals that live there are fascinating—often more bizarre than their depictions in popular culture. Take Finding Nemo’s mum, for example. When she was eaten, in reality, Nemo’s dad would have become his mum so they could continue the family group. On the other hand, Pearl the octopus from the same movie would have 3 hearts and 9 brains, one brain in each arm so that most of her cognitive processing occurred in her detachable but regrowable limbs. She’d also have blue blood because it’s based on copper rather than iron. Or if Patrick the star fish reflected the real world, he’d eject his stomach through his mouth to eat and would likely attempt to eat the entirety of Bikini Bottom, SpongeBob included.
Real life in a coral reef is often stranger than fiction, and sponges don’t disappoint. For example, the deep-sea harp sponge Chondrocladia lyra is stationary but carnivorous, eating fish and crustaceans that swim into their vertical columns (see photos below). In contrast, the tropical sponge Cliona orientalis makes its living as an encrusting sponge, burrowing into a live coral’s skeleton and overgrowing it at the surface.
Though one might mistake them for corals, it’s hard to look around a coral reef and not find an abundance of sponges. In fact, sponges can occupy up to 50% of the reef surface area and, like corals, they make good homes for a wide array of reef organisms (see examples below). More importantly, sponges are critical for the health of coral reefs because they are central to the so-called “Sponge Loop1.” The crux of the sponge loop is this: Although coral reefs are some of the most productive ecosystems on earth, they are often referred to as “marine deserts” because nutrient concentrations in the water column are quite low. Nutrient retention, then, is critical. Sponges filter out large quantities of organic matter (e.g. sugars and proteins) that were released into the water column by organisms living on the reef and convert it into sponge tissue. The sponge cells are then released into the seawater at a high enough rate that the net biomass gain to the sponge is approximately zero. The released sponge cells are then eaten by detritovores, which are then eaten by other organisms that will later release organic matter into the water column, thus completing the loop and keeping nutrients locked within the reef. Although to the human eye sponges appear to be inactive, they are very important components of tropical coral reefs. See here for a demonstration of sponge feeding.

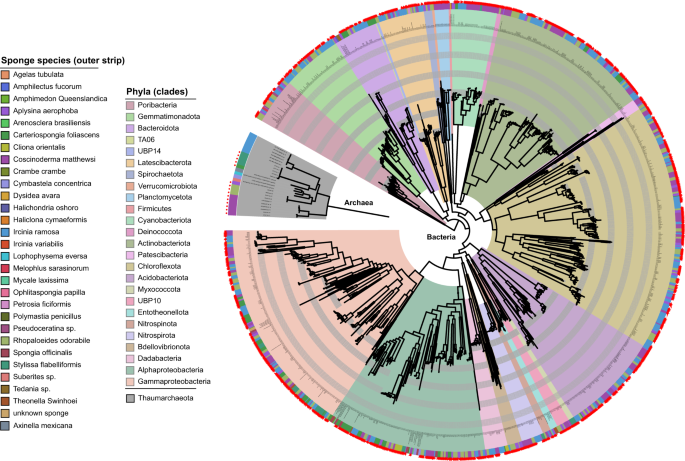
Tropical sponges, like most animals, host a variety of microbial symbionts (bacteria and archaea) that we collectively refer to as the sponge microbiome. These symbionts can make up ~35% of the sponge’s total biomass2 and are thought to carry out a range of processes that are important for maintaining sponge health. For example, sponge symbionts are thought to provide the sponge with vitamins, remove waste ammonia, and potentially participate in the sponge loop by transforming components of dissolved organic matter3. However, these hypotheses are mostly based on 16S rRNA gene surveys that give information on the identities of the symbionts, but a very fuzzy idea of what they might do. A few community-level genome-based characterisations of sponge-symbiont communities have been conducted4,5, but most studies have focussed almost exclusively on specific lineages of interest (e.g. Poribacteria or Chloroflexi6,7). So far, the majority of symbiont lineages found in tropical marine sponges remain largely uncharacterised, hampering our ability to identify core features of sponge symbionts, those that underpin the relationship between the host and its microbes.
To address this, we undertook an analysis of ~1200 metagenome-assembled genomes (MAGs) derived from seven marine sponge species, spanning 25 microbial phyla and the vast majority of microbial taxa commonly found in marine sponges. Interestingly, we identified specific functional guilds for the transformation of sugars common in coral reef dissolve organic matter, suggesting that microbes play a role in the Sponge Loop. This discovery is fascinating as the Sponge Loop has traditionally been described in terms of the sponge itself, while our data suggest that microbes have their roles to play in this important ecological process. We also show that, in contrast to previous reports, the Thaumarchaeota alone (not bacteria) are capable of oxidizing waste ammonia produced by the sponge, and therefore represent a keystone species for this process across sponge taxa. We further found that processes previously thought to be important, such as amino acid provisioning and taurine, creatine, and carnitine metabolism are unlikely to be central mechanisms mediating sponge-microbe symbiosis. Through the work of Willis Song and Torsten Thomas at the University of New South Wales, we also provide evidence that many of the genes thought to underpin sponge-microbe symbiosis, like the glycosyls hydrolases for transforming reef DOM, eukaryote-like proteins for evading the host immune system, and CRISPR and restriction-modification system genes for fending off viral infection, have been laterally transferred between disparate microbial lineages. Fascinatingly, this finding suggests that lateral gene transfer plays an important role in conferring a selective advantage to specific sponge-associated microorganisms. Taken together, these data illustrate how evolutionary processes have partitioned ecological functions across sponge symbiont lineages, allowing them to occupy or share specific niches and live symbiotically with their sponge hosts.
Read the full manuscript here.

1. De Goeij, J. M. et al. Surviving in a marine desert: The sponge loop retains resources within coral reefs. Science (80-. ). 342, 108–110 (2013).
2. Taylor, M. W., Radax, R., Steger, D. & Wagner, M. Sponge-Associated Microorganisms: Evolution, Ecology, and Biotechnological Potential. Microbiol. Mol. Biol. Rev. 71, 295–347 (2007).
3. Pita, L., Rix, L., Slaby, B. M., Franke, A. & Hentschel, U. The sponge holobiont in a changing ocean : from microbes to ecosystems. 6, 1–18 (2018).
4. Slaby, B. M., Hackl, T., Horn, H., Bayer, K. & Hentschel, U. Metagenomic binning of a marine sponge microbiome reveals unity in defense but metabolic specialization. ISME J. 11, 2465–2478 (2017).
5. Engelberts, J. P. et al. Characterization of a sponge microbiome using an integrative genome-centric approach. ISME J. 14, 1100–1110 (2020).
6. Kamke, J. et al. Single-cell genomics reveals complex carbohydrate degradation patterns in poribacterial symbionts of marine sponges. ISME J 7, 2287–2300 (2013).
7. Bayer, K., Jahn, M. T., Slaby, B. M., Moitinho-Silva, L. & Hentschel, U. Marine Sponges as Chloroflexi Hot Spots: Genomic Insights and High-Resolution Visualization of an Abundant and Diverse Symbiotic Clade . mSystems 3, 1–19 (2018).




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in