Vanishing Glaciers: A global-scale project to study the microbiome of glacier-fed streams (GFSs)
We all (should) know the sad truth about glaciers; they are melting down rapidly and probably irreversibly (for a long time at least). This is expected to cause dramatic changes to a long list of related downstream ecosystems, with GFSs being on top of this list. As GFSs are changing, their microbiome -the major life form in this harsh ecosystem- is changing too. And this microbiome is still rather unexplored, despite microbes forming the basis of the streams' food web and orchestrating biogeochemical fluxes that are even of global relevance.
The Vanishing Glaciers project, led by Prof. Tom Battin at EPFL, Switzerland, and financed by The NOMIS Foundation, aims to explore the threatened GFS microbiome now that it is still possible. For that purpose, a diverse team of scientists collects, processes and analyses GFS samples from the remaining glaciers all over the globe.

A first glimpse into the GFS sediment microbiome
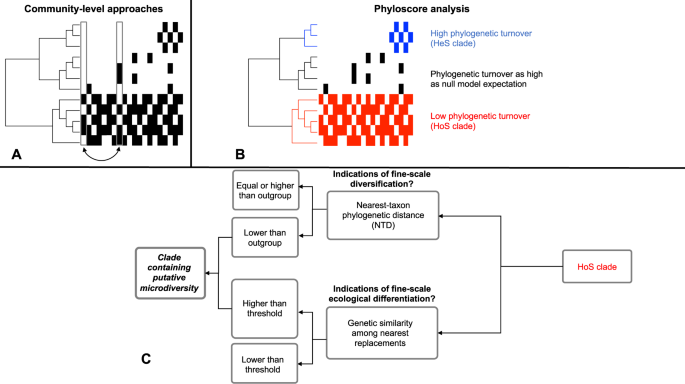
I was lucky enough not just to be part of this team, but also to get my hands on the first chunk of data as it arrived from the first expedition in New Zealand. Our idea here was to go beyond the mere description of the GFS sediment microbiome and to try to understand what ecological mechanisms shape the structure of the microbiome that we observed. Focusing on bacteria and comparing sediment communities within and among the twenty sampled GFSs, we found that almost 90% of the communities are phylogenetically more similar than expected by chance. Phylogenetic similarity usually correlates to functional similarity in the microbial world (at short phylogenetic distances); this means that the examined communities are inferred to be more functionally similar than expected by chance. In microbial ecology this is often attributed to the dominance of "homogeneous selection", i.e., similar "selective environments" governing the structure of the examined communities. In the paper we rephrased it to "homogeneous ecological selection" (HoS) to avoid agitating evolutionary biologists that are sensitive to hearing the s-word (in fact we first agitated them and then we changed it).
The apparent dominance of HoS in the system naturally raised the question of who are the microbial taxa that are driving these patterns. This is important to know because these would represent "ecologically successful" taxa in the sense that they are much more prevalent than others and than expected by chance in the specific ecosystem. (Un)fortunately for us, there was at that time no statistical framework that could identify such taxa, because the existing frameworks examine community-level patterns whereas we wanted to examine patterns for each tip in our phylogenetic tree. So we sat down and thought of how we could develop a new framework to do so. Even though in principle we knew what would be required, it took us quite some time and hardcore R programming to do it properly.
But when we finally finished coding and we applied the code to the dataset from New Zealand, the results were worth the effort; we had for the first time an idea of the ecologically successful phylogenetic clades in GFS sediments. The identified clades, which we called "HoS clades", contained more than 40% of the total observed bacterial α-diversity and two out of three bacterial cells per gram of sediment. They included groups with a wide metabolic repertoire that often involves carbon fixation via chemolithoautotrophy, and they were indeed even more abundant and prevalent in samples with lower levels of primary production (i.e., chl-a concentration).
In nature, bacterial groups with broad spatial distributions like our HoS clades often achieve that via microdiversity, i.e., ecological differentiation between genetically very similar taxa. Because of that, we also examined our HoS clades for signs of microdiversity. Indeed, we observed that despite being more prevalent in GFS sediments, taxa in HoS clades were much more similar phylogenetically compared to the rest of the microbiome. Moreover, spatial "replacements" (related to ecological differentiation) occurred among more genetically similar taxa in HoS clades than in the rest of the microbiome. These are two solid signs of microdiversity in HoS clades, which we however still call "putative" microdiversity in the sense that we did not experimentally quantify the degree of ecological distinctness of closely related taxa.
The time to act is now
Despite the fact that we learned a lot more about the important clades in the GFS sediment microbiome, our findings made us realise that these clades are probably in big trouble. This is because the environment of GFSs is predicted to change dramatically as glaciers shrink owing to climate change. As turbidity decreases in GFSs due to reduced discharge and sediment loads, the environment will become more advantageous for primary production and this is bad news for our HoS clades that don't seem to get along well with primary producers. This urges towards in-depth characterisation of these clades whose putative microdiversity could even be translated into unexploited potential for biotechnology.
We are continuing the efforts from our side, sampling more and more glacier-fed streams all around the world with a target of having ~200 sampled by the end of 2022. And we are gathering data from multiple aspects to characterise their microbiome, including shotgun metagenomics and enzymatic activities on top of 16S rRNA and 18S rRNA gene amplicon sequencing. Stay tuned!
Links related to the Vanishing Glaciers project
Project homepage: https://www.glacierstreams.ch/
The project at The NOMIS Foundation website: https://nomisfoundation.ch/research-projects/vanishing-glaciers-what-else-besides-water-is-lost/
Expedition blog: https://epfloutthere.tumblr.com/tagged/vanishingglaciers




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in