Bacteria use a variety of mechanisms to compete with neighbouring bacterial cells1. A widespread pathway for interbacterial antagonism is the type VI secretion system (T6SS), which injects toxic antibacterial effector proteins into target cells2. In our recent work (https://www.nature.com/articles/s41586-019-1735-9), we characterize a novel T6SS effector exported by the opportunistic pathogen Pseudomonas aeruginosa.
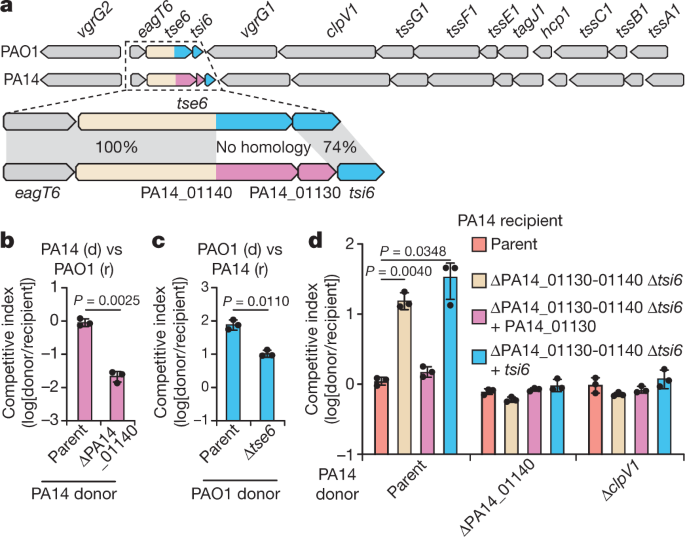
The toxin domains of T6SS effectors are typically found at their C-terminus. Shortly after beginning my PhD studies at McMaster University, I made the observation that one of the six well-characterized T6SS effectors of P. aeruginosa possessed no homology at its C-terminus between the model strains PAO1 and PA14 (Fig. 1). In PAO1, this effector is named Tse6 and encodes an NAD(P)+ glycohydrolase toxin, whereas, in the strain PA14, the predicted toxin domain appeared to be unique. T6SS effectors are encoded adjacent to immunity genes, which encode for proteins that neutralize their cognate toxins. The uncharacterized PA14 toxin had an immunity gene that was distinct from that found in PAO1, further suggesting that it possessed novel activity.

Figure 1. Comparison between the tse6-tsi6 and the PA14_01140-PA14_01130 effector-immunity pairs from P. aeruginosa strains PAO1 and PA14, respectively. Τhe known toxin-immunity-encoding regions of tse6-tsi6 and the predicted toxin-immunity-encoding regions of PA14_01140-PA14_01130 are shown in blue and pink, respectively. Genes encoding T6SS components are shown in gray.
After demonstrating that the PA14 toxin inhibited the growth of E. coli and P. aeruginosa, we used a structural biology approach to understand more about how it kills bacteria. With the help of our colleague Peter Stogios at the Center for Structural Genomics of Infectious Disease in Toronto, we were able to solve the crystal structure of the PA14 toxin bound to its immunity protein. Analysis of this structure revealed that our toxin resembled RelA-SpoT homolog proteins that synthesize a molecule called (p)ppGpp, an essential signaling nucleotide in bacteria that mediates survival under nutrient-limiting conditions3. This made perfect sense to us because (p)ppGpp signaling is known to drive cells into a state of dormancy, which would be a reasonable mechanism for growth inhibition by an interbacterial toxin. However, my initial attempts to detect (p)ppGpp produced by the PA14 toxin were unsuccessful. I could not generate chromatograms typical of (p)ppGpp synthetases and could not figure out why.
Around the same time as I was trying to figure out the biochemical activity of my toxin, I went to the 2018 Gordon Research Conference on Microbial Stress Response. It was here that I met Dr. Boyuan Wang, a postdoc in Michael Laub’s lab at the Massachusetts Institute of Technology. At the time, Boyuan’s postdoctoral work was heavily focused on (p)ppGpp biochemistry and so after explaining my project to him in detail, we initiated a collaboration to determine the activity of our toxin.
Enzymes that synthesize (p)ppGpp use one molecule of ATP to donate a pyrophosphate group to either a GDP or GTP acceptor, forming ppGpp or pppGpp, respectively4. After shipping Boyuan our purified toxin, he determined that it was unable to pyrophosphorylate guanine nucleotides as was suggested by our crystal structure. Instead, he found that it specifically pyrophosphorylated ADP and ATP, producing the poorly characterized nucleotides ppApp and pppApp, respectively. An enzyme that synthesizes (p)ppApp had not been previously characterized and for this reason, we renamed the toxin to type VI (p)ppApp synthetase 1 (Tas1).
We soon after found that Tas1 is able to produce (p)ppApp at rates of 180,000 molecules per minute in vitro. Using metabolite profiling, we determined that Tas1 expression in P. aeruginosa and E. coli cells results in the rapid production of (p)ppApp and concomitant depletion of ADP and ATP, which substantially alters the cellular metabolome and causes cell death.
Because (p)ppApp is structurally similar to (p)ppGpp we also wondered if in addition to depleting essential adenosine nucleotides, (p)ppApp could also act as a ligand for any of the known (p)ppGpp targets in the bacterial cell. In early 2019, I visited the Laub lab to learn from and work with Boyuan to address this idea. During this time, we focused on PurF, a purine biosynthesis enzyme that Boyuan had previously found to be inhibited by (p)ppGpp5. We were able to show that ppApp similarly binds and inhibits the activity of PurF suggesting that the accumulation of (p)ppApp not only results in a collapse of the metabolic output of the cell, but also results in inhibition of purine biosynthesis. Therefore, Tas1 likely uses a dual-pronged attack to not only deplete cells of ATP but also inhibit synthesis of this nucleotide to prevent cell recovery (Fig. 2).

Figure 2. Tas1 likely uses a dual-pronged attack upon entering target cells. Attacking cells (blue) inject Tas1 into recipient cells (orange) using their T6SS (green). This causes rapid depletion of ADP and ATP through the production of (p)ppApp (1), which inhibits the purine biosynthesis enzyme PurF (2). Inhibition of PurF prevents the synthesis of 5’-phosphoribosylamine (5’-PRA) from phosphoribosyl pyrophosphate (PRPP) and hinders the production of adenosine and guanosine nucleotides. Together, this results in dysregulation of the cellular metabolome and prevents recipient cell recovery by inhibiting ATP production (3).
In this work, we show that Tas1, despite resembling (p)ppGpp synthetases, depletes cellular ATP by rapidly synthesizing (p)ppApp. This work has initiated several questions about the prevalence of (p)ppApp producing enzymes. The identification of additional (p)ppApp synthetases may allow us to identify roles for these enzymes that extend beyond toxicity and instead, uncover novel cell signaling pathways that are distinct from those of (p)ppGpp.
References:
- Granato, E. T., Meiller-Legrand, T. A. & Foster, K. R. The evolution and ecology of bacterial warfare. Curr. Biol. 29, R521–R537 (2019).
- Russell, A. B., Peterson, S. B. & Mougous, J. D. Type VI secretion system effectors: poisons with a purpose. Nat. Rev. Microbiol. 12, 137–148 (2014).
- Hauryliuk, V., Atkinson, G. C., Murakami, K. S., Tenson, T. & Gerdes, K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol. 13, 298–309 (2015).
- Atkinson, G. C., Tenson, T. & Hauryliuk, V. The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One 6, e23479 (2011).
- Wang, B. et al. Affinity-based capture and identification of protein effectors of the growth regulator ppGpp. Nat. Chem. Biol. 15, 141–150 (2019).





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in