Many human genetic targets are not readily ‘druggable’ with conventional strategies, but they are potentially druggable with gene therapy, which enables gene replacement or silencing to address disease-related mutations (Deverman et al., 2018). Adeno-associated viral (AAV) vectors have emerged as leading gene delivery tools for therapeutic applications (Kotterman and Schaffer, 2014; Zinn and Vandenberghe, 2014). Together with Luxturna was approved by the FDA in 2017 for the treatment of retinitis pigmentosa (Dias et al., 2018), a set of AAV-based gene therapies are in clinical research.
Optimization of AAV vectors will benefit its therapeutic usages. For example, in 2016, novel, engineered AAV capsids were reported to have >40‑fold advancement to transfer genes to the CNS in the adult mouse compared with the previous standard AAV9, potentially transforming the ability to treat neurodegenerative diseases with AAV gene therapy (Deverman et al., 2016). The knowledge of how AAV interacts with host cellular factors to facilitate infection and how AAV receptors contribute to AAV vector transduction efficiency and tropism is demanded for further optimization of AAV vectors. In 2016, adeno-associated virus receptor (AAVR), which is a glycosylated protein containing 5 polycystic kidney disease (PKD) repeat domains, was identified as the essential receptor of multiple AAV serotypes (Pillay et al., 2016). Different AAV serotypes have evolved distinctive interactions with the same receptor AAVR (Pillay et al., 2016; Pillay et al., 2017), AAV2 (clade B) predominantly interacts with PKD2, AAV5 (no designated clade) interacts primarily through PKD1, while other AAV serotypes, including AAV1 (clade A) and AAV8 (clade E), require a combination of PKD1 and PKD2 for efficient transduction (Pillay et al., 2016; Pillay et al., 2017).
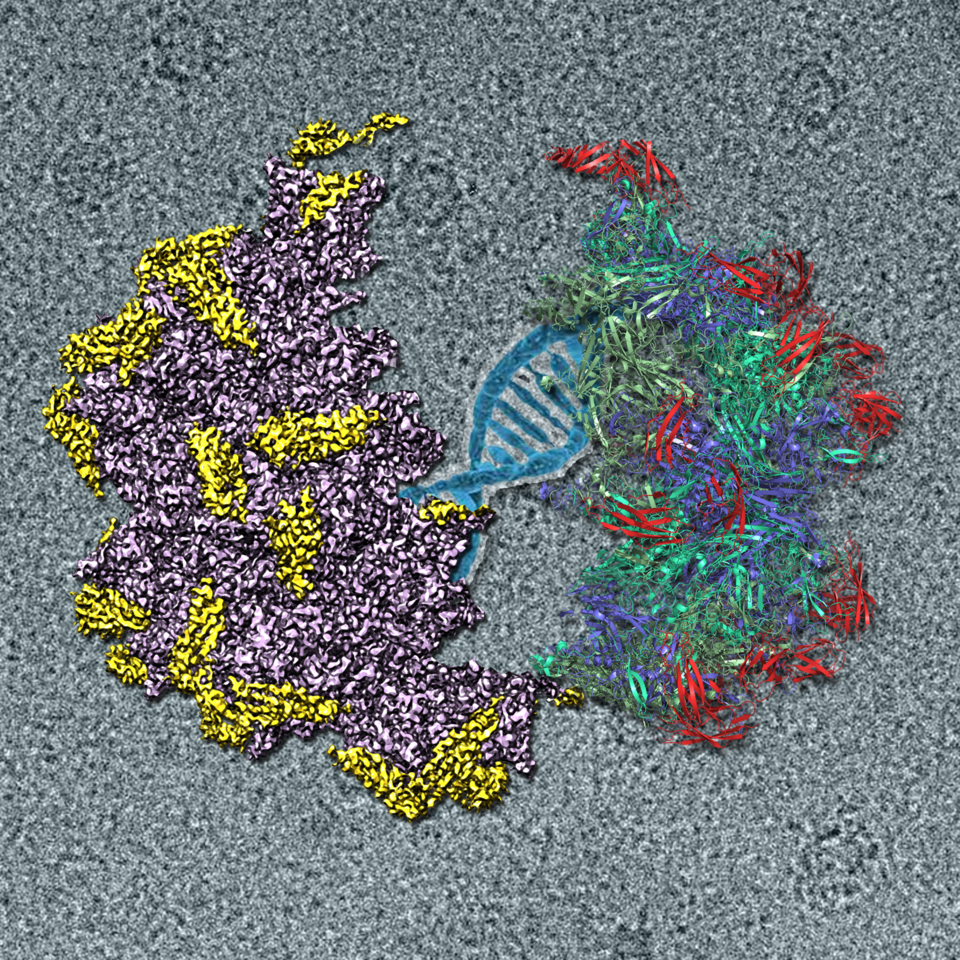
As AAV2 is the most widely used vector in gene therapy, we recently report the cryo-EM structure of the AAV2-AAVR complex at 2.8 Å resolution. This structure reveals that of the five Ig-like PKDs in AAVR, PKD2 binds directly to the spike region of the AAV2 capsid adjacent to the icosahedral 3-fold axis. Residues in strands B and E, and the BC loop of AAVR PKD2 interact directly with the AAV2 capsid. Interestingly, the interacting residues in the AAV2 capsid are mainly in AAV-featured variable regions (VRs). The sole engagement between AAV2 and AAVR PKD2 is well fitted with the previous functional research (Pillay et al., 2016; Pillay et al., 2017). As the key interacting residues on AAV2-AAVR significantly affect virus attachment and infectivity, we hope our structural results will be helpful to structure-based rational design of AAV2 to increase virus-receptor binding ability and virus infectivity.

But there are still several key questions need to be answered. For example, AAVR engages multiple AAV serotypes with various rules. What is the molecular mechanism of the divergent rules? Why AAVR contacts AAV serotypes with various strategies? We hope we could answer these questions in the future. Our report can be found at https://www.nature.com/articles/s41564-018-0356-7 .




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in