Written by Mike Ellis and Caressa Tsai
There exists a paradox in modern antibiotic discovery. Antibacterial efficacy is routinely assessed against bacteria grown in standard microbiological media (e.g. Mueller-Hinton Broth, MHB), despite a wealth of evidence that the genetic requirements for bacterial growth during infection are vastly different from those in nutrient-rich growth media. Our paper published yesterday in Nature Communications describes an approach to find novel antibacterials by screening a chemical library against Salmonella Typhimurium grown in conditions that closely mimic the in vivo environment.
We began this project in 2017 as new trainees in the labs of Brian Coombes (Caressa, PhD student) and Eric Brown (Mike, post-doc). At the beginning, the project goal was straightforward: identify compounds that have antibacterial activity against Salmonella Typhimurium grown in infection-relevant conditions. Looking back, we wouldn’t have been successful if it weren’t for the unique expertise from our respective labs (Coombes, in vitro and in vivo models for Salmonella pathogenesis; Brown, high-throughput compound screening and chemical genomics for antibiotic discovery).
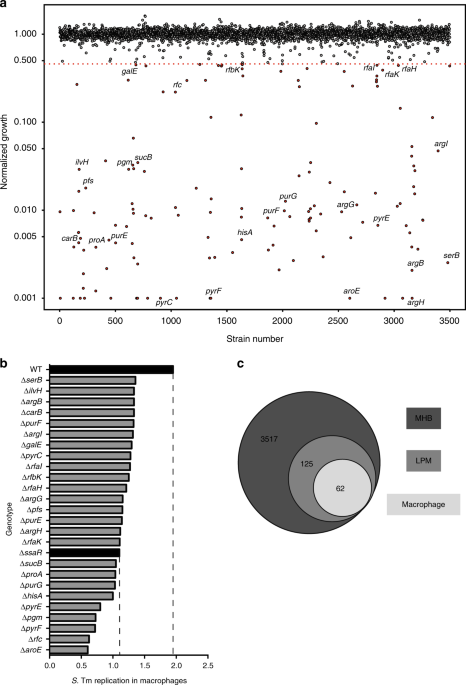
In May 2017 we conducted four chemical screens with a small library of ~4,000 molecules against Salmonella grown in the presence of sub-inhibitory concentrations of hydrogen peroxide, bile salts, a defined media designed to mimic the intracellular environment (LPM), and standard nutrient-rich growth media (MHB). As we anticipated, the presence of low amounts of peroxide or bile salts sensitized WT Salmonella to killing by a total of ~70 compounds that had no activity in MHB. However, we were surprised to observe that growth in LPM sensitized Salmonella to ~150 compounds that had no antibacterial activity in the any of the three other screens. This experiment (not included in our publication) was our first hint that the intracellular environment might induce remarkable antibiotic sensitivity, and led us to focus on identifying compounds with selective activity against Salmonella after internalization into phagocytic cells.
Like several other intracellular pathogens, the ability for Salmonella to survive and replicate within host phagocytes is essential for virulence in vivo. Much of our current understanding of Salmonella’s intracellular lifestyle has been derived from gentamicin protection assays, in which macrophages cultured with Salmonella are lysed and plated to enumerate bacterial load at specific time points. With the addition of a constitutive luciferase reporter system and a few other modifications, Caressa adapted this routinely used assay into one amenable for high-throughput screening. By September 2017, we were screening a library of 1,600 previously approved drugs against intracellular Salmonella alongside screens in LPM and MHB. We were excited to find a number of compounds that were effective against intracellular Salmonella with only modest activity against bacteria grown in LPM, and no measurable activity in MHB.
We initially encountered some difficulty in reconciling the discrepancy between compound activity against Salmonella grown in either macrophages or LPM, despite the intracellular-mimicking nature of the latter. However, Mike considered the potential impact of antimicrobial peptides present within host cells, and therefore did a chequerboard assay with an antimicrobial peptide (polymyxin B) and one of our most promising leads (metergoline). Excitingly, the addition of this membrane-perturbing agent allowed us to observe antibacterial activity against Salmonella grown even in nutrient-rich microbiological media (MHB). By January 2018, Caressa had shown that metergoline worked in vivo to extend animal survival during a lethal Salmonella infection, and Mike had found that chemical or genetic perturbation of the outer membrane was sufficient for metergoline to work in MHB.

Over the next few months we continued to characterize the activity of metergoline with several exciting in vitro and in vivo experiments. During this time, we were also fortunate to initiate a helpful collaboration with Jake Magolan, who had just moved his medicinal chemistry lab to our research institute at McMaster University. After a short chat about our project, just two weeks later Jake’s post-doc (Jarrod Johnson) had synthesized the first batch of metergoline analogues for us to test, offering a promising starting point to enhance its modest in vivo activity through optimization of the chemical scaffold.
Ultimately, we identified metergoline as a novel small molecule with antibacterial activity. Along the way, we were also able to gather evidence to suggest that the Salmonella outer membrane is disrupted in the intracellular environment, inducing sensitivity to molecules that are otherwise unexpected to possess activity against Gram-negative bacteria. We attribute this to Mg2+ depletion in the Salmonella-containing vacuole, and offer a model to provide a parsimonious explanation for the differences in antibacterial activity against bacteria grown in infection-relevant conditions. Looking forward, we envision that future screens against intracellular Salmonella will identify new antibiotic leads against Gram-negative bacteria. Our work suggests that the Gram-negative outer membrane is not an effective permeability barrier in this environment, so we encourage others to test any new Gram-positive targeting antibacterials against intracellular Gram-negative bacteria.






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in