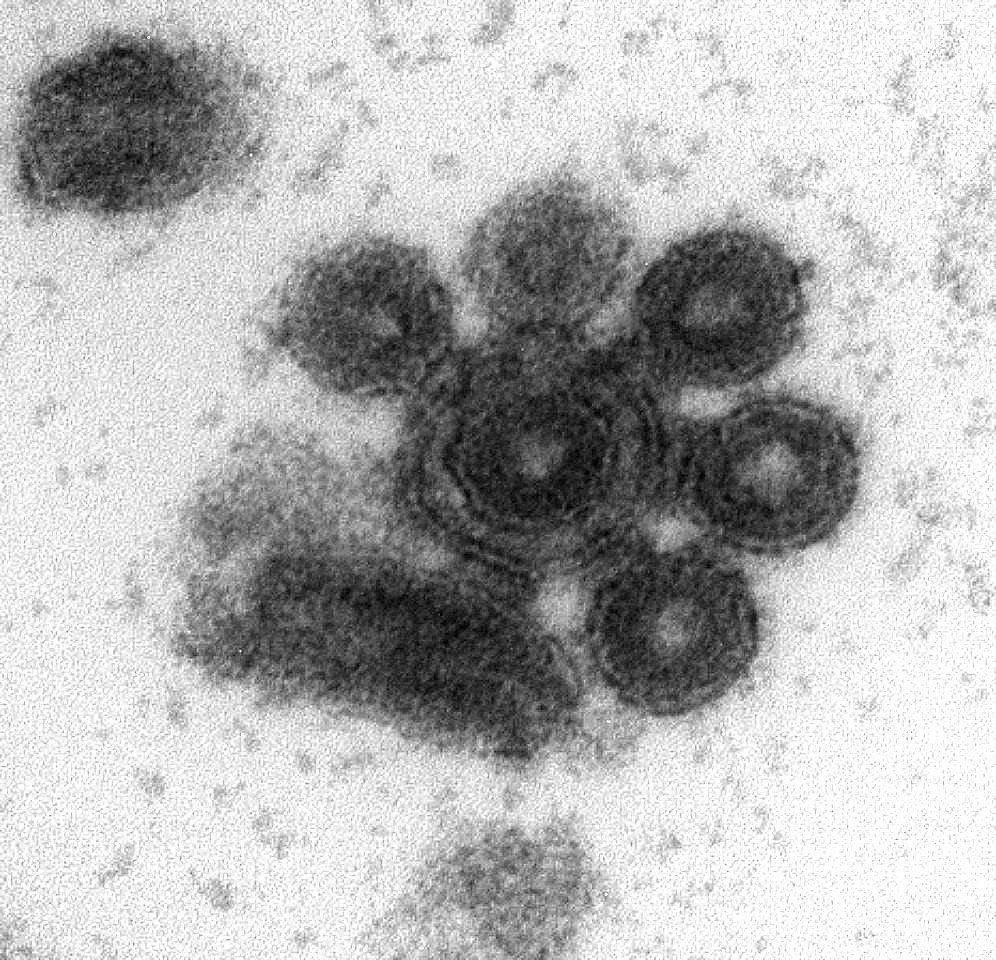
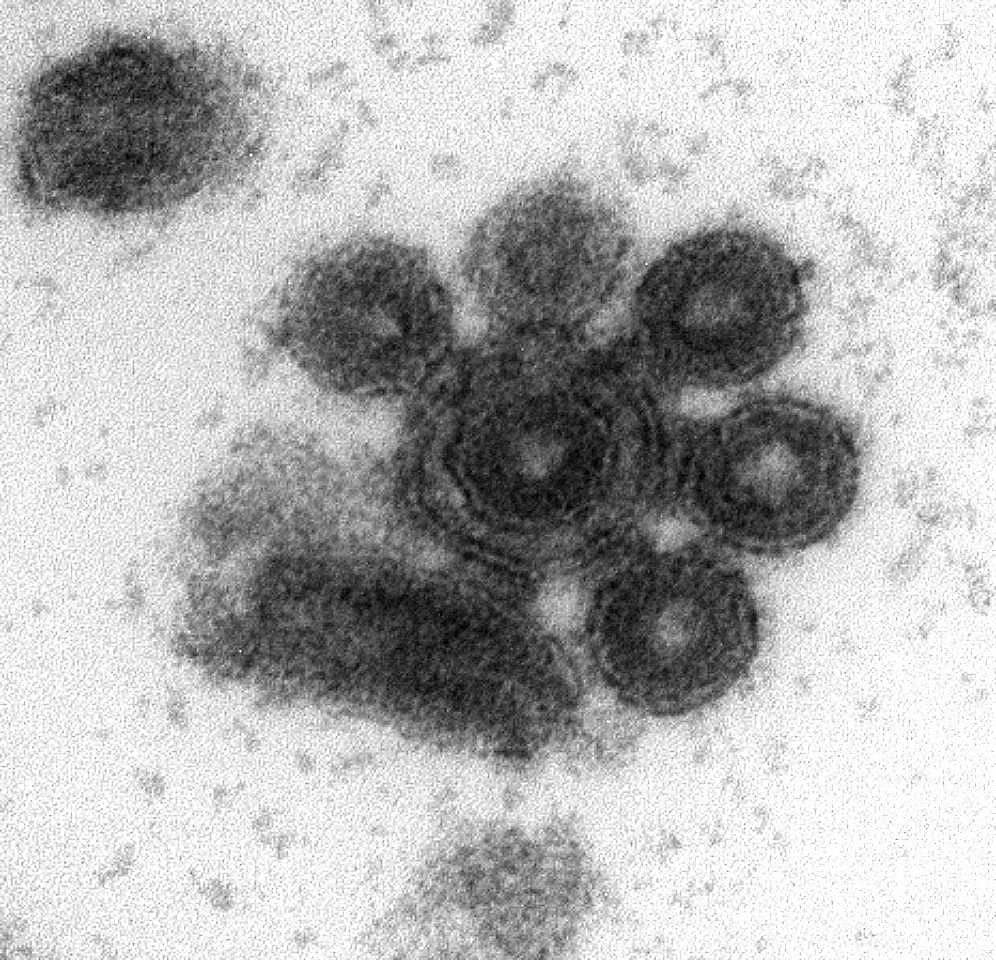
Early electron microscopy studies found that viral particles could form aggregates in a variety of viruses including tobacco mosaic virus, poxviruses, influenza virus, vesicular stomatitis virus (VSV), and poliovirus. However, the molecular basis and implications of these observations have remained largely unexplored. Recently, we analyzed the VSV genetic diversity produced by single infected cells, and surprisingly, we found that single infectious units were sometimes constituted by multiple viral genomes1. This finding led us to explore more in depth the biological implications of viral aggregation.
Many animal viruses, such as VSV, are enveloped in a lipid bilayer uptaken from cellular membranes. Since viral surface proteins bind to these membranes to initiate infection, we hypothesized that free virions may also be capable of interacting with the envelopes of other virions extracellularly.
Using microscopy, dynamic light scattering, differential centrifugation, and flow cytometry, we have shown here that free viral particles can spontaneously aggregate into multi-virion infectious units. Following the establishment of these contacts, different viral genetic variants can be co-transmitted to the same target cell. This process can be enforced, such that a “crippled” virus unable to enter cells on its own can be assisted by another, fully functional virus, and complete its infection cycle successfully. In purified virions, this process is driven by protein-lipid interactions probably involving the VSV surface glycoprotein and phosphatidylserine.
Since the oral cavity is a major site of infection for VSV, causing stomatitis, we set out to test for virion aggregation in saliva, initially from humans and later on, from cows (a more frequent VSV host). We were excited about this kind of “field work”, which is so different from our daily lab routine. We found that multi-virion complexes were very abundant in both human and cow saliva. This is particularly relevant, since the saliva is an important route of viral shedding and could act as a vehicle for viral transmission. However, aggregation was highly variable depending on the subject, for reasons that we still remain to be investigated. It also remains to be shown whether the mechanism of aggregation is the same as in purified virions or differs.
Our findings contrast with the commonly accepted perception of virions as passive propagules, and show the ability of enveloped viruses to establish “collective infectious units”, which could in turn facilitate the evolution of virus-virus interactions and of social-like traits. This is now becoming a hot topic, and a highly related paper using very similar techniques appeared during the peer review process of our work. This related paper suggests that aggregation may also play an important role in the biology of poliovirus2. Those interested in further reading can also check a recent review article by our senior author3.
The paper in Nature Microbiology is here: http://go.nature.com/2rcWKYk
- Combe,M., Garijo,R., Geller,R., Cuevas,J.M. & Sanjuan,R. Single-Cell Analysis of RNA Virus Infection Identifies Multiple Genetically Diverse Viral Genomes within Single Infectious Units. Cell Host Microbe. 18, 424-432 (2015).
- Aguilera,E.R., Erickson,A.K., Jesudhasan,P.R., Robinson,C.M. & Pfeiffer,J.K. Plaques Formed by Mutagenized Viral Populations Have Elevated Coinfection Frequencies. MBio. 8, e02020-16 (2017).
- Sanjuán, R. Collective Infecitous Units in Viruses. Trends Microbiol, in press. doi: 10.1016/j.tim.2017.02.003



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in