One of the biggest challenges facing the growing field of 'viromics' or 'phageomics' is the difficulty in assigning bacterial hosts to newly discovered in silico phage (bacteriophage genomes assembled from individual reads from metagenomic sequencing projects). There are some bioinformatic hints as to the likely host, but these are not very satisfactory. Also of course, to properly study a phage, you need to be able to grow it in the lab, for which you need a susceptible host. Here I describe our successful strategy to breathe life into crAssphage - the most abundant member of the human gut virome.
__
At APC Microbiome Ireland we have been studying the gut microbiome for over 15 years. We entered the field at a time when only 100 papers a year were being published on the human microbiome, little realising the explosion in microbiome research that was about to occur (4672 papers in 2017 - Web of Science). Of course, like many others, most of our studies focused on the ‘bacteriome’. Only recently Paul Ross and I have become interested in the virome, sometimes referred to as the ‘dark matter’ of the microbiome. Among the questions we asked were what role phage may play in microbiome structure, whether or not they have a direct effect on human physiology, and whether we can use them as biomarkers of microbiome stability and diversity?
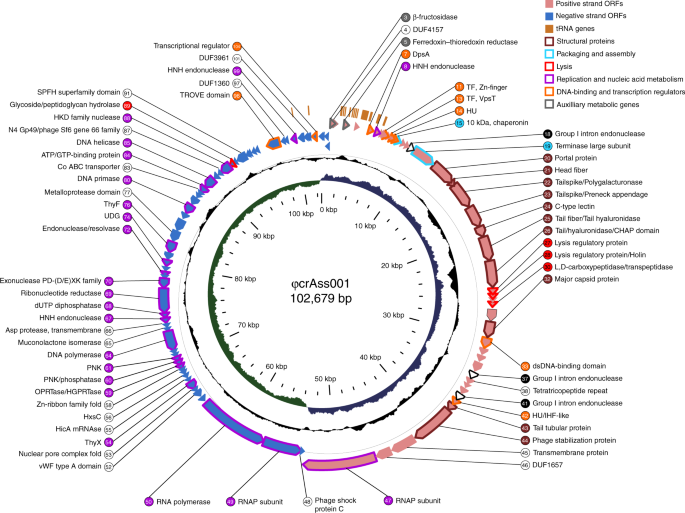
One of the things that immediately captured our attention on entering the field was the extraordinary discovery of crAssphage by Bas Dutilh, Rob Edwards and colleagues (published in Nature Communications in 2014). They showed that this highly abundant genome could be assembled from the sequencing reads of multiple individuals and then from one individual (crAss - cross Assembly). This predicted 97kb bacteriophage had no known host, but represented up to 90% of all the bacteriophage sequences in some individuals. This looked like an amazing challenge for our fledgling group. Could we isolate and biologically characterise the most abundant representative in the human virome? What does crAssphage look like, what does it kill, what kind of replication strategy could support such an abundance of phage without wiping out its host?
So, we set out to see whether or not we could play the role of the Blue Fairy in Pinnochio and turn an in silico phage into a ‘real’ phage? Andrey Shkoporov and Ekaterina Khokhlova, two outstanding scientists in the group, were willing take on this daunting task, ably supported by Brian Fitzgerald, Stephen Stockdale and Lorraine Draper. The key breakthrough (which fittingly has just been published in Nature Communications) came when we isolated 54 strains from the gut microbiome and grew them in pure culture. Then we added the collected viromes from six individuals to each purified strain and sub-cultured three times, followed by metagenomic sequencing and assembly. Lo and behold, in one instance we saw a massive expansion of a familiar looking sequence – crAssphage! The host turned out to be a strain of Bacteroides intestinalis, validating the prediction by Bas and Rob that the host was likely to be a species of Bacteroides.
Of course, now we have a ‘real’ phage and a susceptible host, and so the really interesting work has begun. We showed that crAss001 can replicate in the presence of its host without limiting host growth, we performed proteomics on the isolated phage particles, annotated the genome, and we confirmed by electron microscopy that is a podovirus - and a particularly beautiful specimen at that. We are confident that this approach will also work with other crAssphage, and other phage currently known only by their sequences, and we look forward to breathing life into many more in silico phage over the coming years.




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in
This is the link to the research article:
ΦCrAss001 represents the most abundant bacteriophage family in the human gut and infects Bacteroides intestinalis
https://www.nature.com/articles/s41467-018-07225-7